 Research at the UKE?
Research at the UKE?
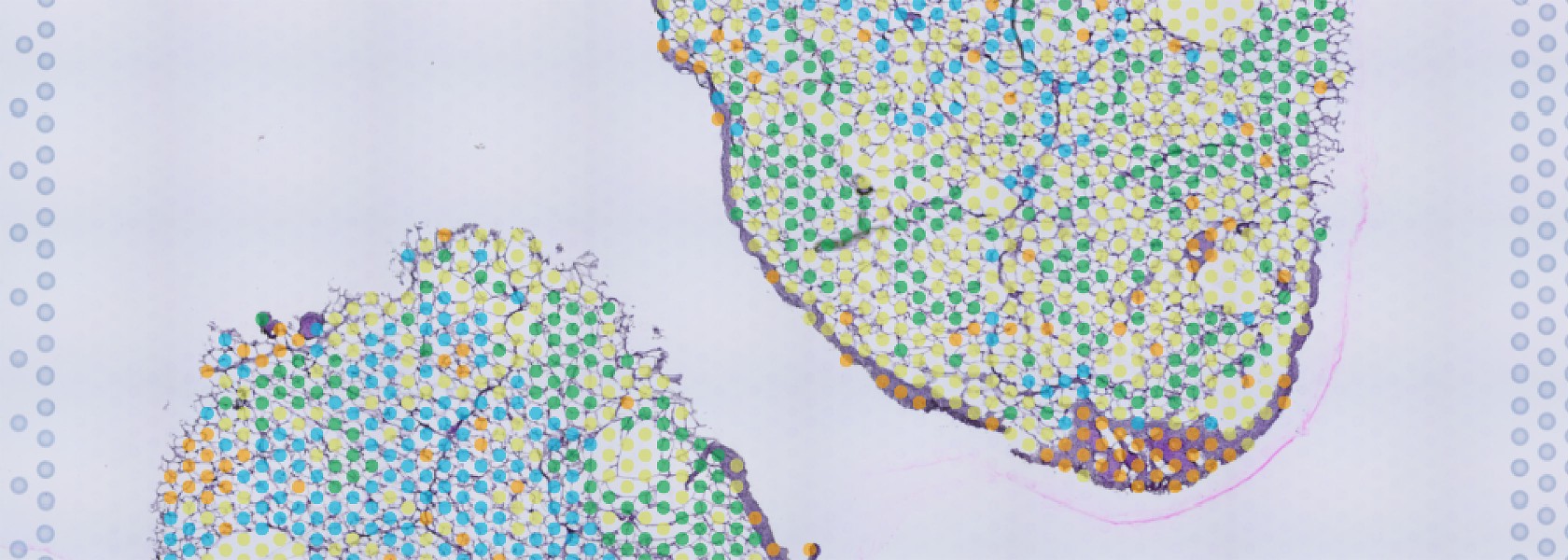
Adlung Lab – Adipose Systems Immunology
The Adlung Lab addresses the unmet need for a systems-level understanding of adipose tissue changes as obesity and its co-morbidities increase worldwide. Our interdisciplinary efforts combine deep molecular profiling with mathematical modelling, ultimately translating into novel therapies for diseases ranging from type 2 diabetes to cancer.

“I want to understand how adipose tissue changes over time during disease development. Mechanistic understanding is a prerequisite for rational intervention.”
Dr. Lorenz Adlung
Project details and goals
Lipid-Associated Macrophage (LAM) cells play a central role in immune-metabolic regulation. However, their role as an anti-inflammatory therapeutic in adipose tissue and beyond remains to be explored.
We are studying adipose systems immunology to:
- ameliorate obesity-associated inflammation with LAM cell-based interventions
- elucidate the cross-talk between immune cells, adipocytes and stromal cells
- derive mechanistic insight into molecular and cellular dynamics during inflammation
Current projects – Adlung Lab

Towards LAM cell-based anti-inflammatory interventions
We have developed a statistical model that integrates transcriptome- and proteome-wide information to understand the mechanism by which LAM cells acquire their lipid handling and anti-inflammatory phenotype. The model predicts that macrophages generated in vitro enhance phagocytosis, lipid processing and oxidative phosphorylation, which we functionally validate. We show that the molecular processes of in vitro generated LAM cells match key functions of their in vivo counterparts.

LAM/adipocyte-ratio inference for rational interventions into obesity
The idea is to dissect differential gene expression programmes from LAM cells and adipocytes, which are present in different ratios with high variability between individuals. The variability in the LAM/adipocyte ratio can (only) partially be explained by the grade of adiposity and therefore requires an interdisciplinary approach.

scMod: From single-cell data to immune dynamics
In this project, we aim to predict how the immune system responds over time to external cues such as tissue damage, infection or tumour formation. Our predictions are based on snapshot information from single-cell data such as flow cytometry or mRNA sequencing. We validate computer simulations with targeted molecular measurements.
Team – Adlung Lab


Dr. Lorenz Adlung
Head
E-mail address:




Key Publications – Adlung Lab
Kilian C*, Ulrich H*, Zouboulis V, Sprezyna P, Schreiber J, Landsberger T, Büttner M, Biton M, Villablanca EJ, Huber S, Adlung L. Longitudinal single-cell data informs deterministic modelling of inflammatory bowel disease. NPJ Syst Biol Appl. Accepted. doi:
Abstract:
Single-cell-based methods such as flow cytometry or single-cell mRNA sequencing (scRNA-seq) allow deep molecular and cellular profiling of immunological processes. Despite their high throughput, however, these measurements represent only a snapshot in time. Here, we explore how longitudinal single-cell-based datasets can be used for deterministic ordinary differential equation (ODE)-based modelling to mechanistically describe immune dynamics. We derived longitudinal changes in cell numbers of colonic cell types during inflammatory bowel disease (IBD) from flow cytometry and scRNA-seq data of murine colitis using ODE-based models. Our mathematical model generalised well across different protocols and experimental techniques, and we hypothesised that the estimated model parameters reflect biological processes. We validated this prediction of cellular turnover rates with KI-67 staining and with gene expression information from the scRNA-seq data not used for model fitting. Finally, we tested the translational relevance of the mathematical model by deconvolution of longitudinal bulk mRNA-sequencing data from a cohort of human IBD patients treated with olamkicept. We found that neutrophil depletion may contribute to IBD patients entering remission. The predictive power of IBD deterministic modelling highlights its potential to advance our understanding of immune dynamics in health and disease.
Adlung L*, Cohen Y*, Mor U*, Elinav E. Machine learning in clinical decision making. Med. 2021 Jun 11;2(6):642-665.
Abstract:
Machine learning is increasingly integrated into clinical practice, with applications ranging from pre-clinical data processing, bedside diagnosis assistance, patient stratification, treatment decision making, and early warning as part of primary and secondary prevention. However, a multitude of technological, medical, and ethical considerations are critical in machine-learning utilization, including the necessity for careful validation of machine-learning-based technologies in real-life contexts, unbiased evaluation of benefits and risks, and avoidance of technological over-dependence and associated loss of clinical, ethical, and social-related decision-making capacities. Other challenges include the need for careful benchmarking and external validations, dissemination of end-user knowledge from computational experts to field users, and responsible code and data sharing, enabling transparent assessment of pipelines. In this review, we highlight key promises and achievements in integration of machine-learning platforms into clinical medicine while highlighting limitations, pitfalls, and challenges toward enhanced integration of learning systems into the medical realm.
Adlung L*, Stapor P*, Tönsing C*, Schmiester L, Schwarzmüller LE, Postawa L, Wang D, Timmer J#, Klingmüller U#, Hasenauer J, Schilling M. Cell-to-cell variability in JAK2/STAT5 pathway components and cytoplasmic volumes defines survival threshold in erythroid progenitor cells. Cell Rep. 2021 Aug 10;36(6):109507.
Abstract:
Survival or apoptosis is a binary decision in individual cells. However, at the cell-population level, a graded increase in survival of colony-forming unit-erythroid (CFU-E) cells is observed upon stimulation with erythropoietin (Epo). To identify components of Janus kinase 2/signal transducer and activator of transcription 5 (JAK2/STAT5) signal transduction that contribute to the graded population response, we extended a cell-population-level model calibrated with experimental data to study the behavior in single cells. The single-cell model shows that the high cell-to-cell variability in nuclear phosphorylated STAT5 is caused by variability in the amount of Epo receptor (EpoR):JAK2 complexes and of SHP1, as well as the extent of nuclear import because of the large variance in the cytoplasmic volume of CFU-E cells. 24-118 pSTAT5 molecules in the nucleus for 120 min are sufficient to ensure cell survival. Thus, variability in membrane-associated processes is sufficient to convert a switch-like behavior at the single-cell level to a graded population-level response.
Jaitin DA*, Adlung L*, Thaiss CA*, Weiner A*, Li B*, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, Keren-Shaul H, David E, Zmora N, Eldar SM, Subezky N, Shibolet O, Hill DA, Lazar MA, Colonna M, Ginhoux F, Shapiro H, Elinav E, Amit I. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell. 2019 Jul 25;178(3):686-698.e14.
Abstract:
Immune cells residing in white adipose tissue have been highlighted as important factors contributing to the pathogenesis of metabolic diseases, but the molecular regulators that drive adipose tissue immune cell remodeling during obesity remain largely unknown. Using index and transcriptional single-cell sorting, we comprehensively map all adipose tissue immune populations in both mice and humans during obesity. We describe a novel and conserved Trem2+ lipid-associated macrophage (LAM) subset and identify markers, spatial localization, origin, and functional pathways associated with these cells. Genetic ablation of Trem2 in mice globally inhibits the downstream molecular LAM program, leading to adipocyte hypertrophy as well as systemic hypercholesterolemia, body fat accumulation, and glucose intolerance. These findings identify Trem2 signaling as a major pathway by which macrophages respond to loss of tissue-level lipid homeostasis, highlighting Trem2 as a key sensor of metabolic pathologies across multiple tissues and a potential therapeutic target in metabolic diseases.
Alumni - PhD | MD
- NA.