 Research at the UKE?
Research at the UKE?
Bonn Lab – Systems Biology
At the Institute of Medical Systems Biology (IMSB) we are dedicated to unraveling the complexities of human pathology, by combining expertise in AI, machine learning, bioinformatics, and image analysis. Currently, we are focusing on understanding cell to cell signaling in immune mediated and other diseases.

“We enhance understanding of human pathology with computational methods, improving patient outcomes through innovative systems.”
Prof. Dr. Stefan Bonn
Project details and goals
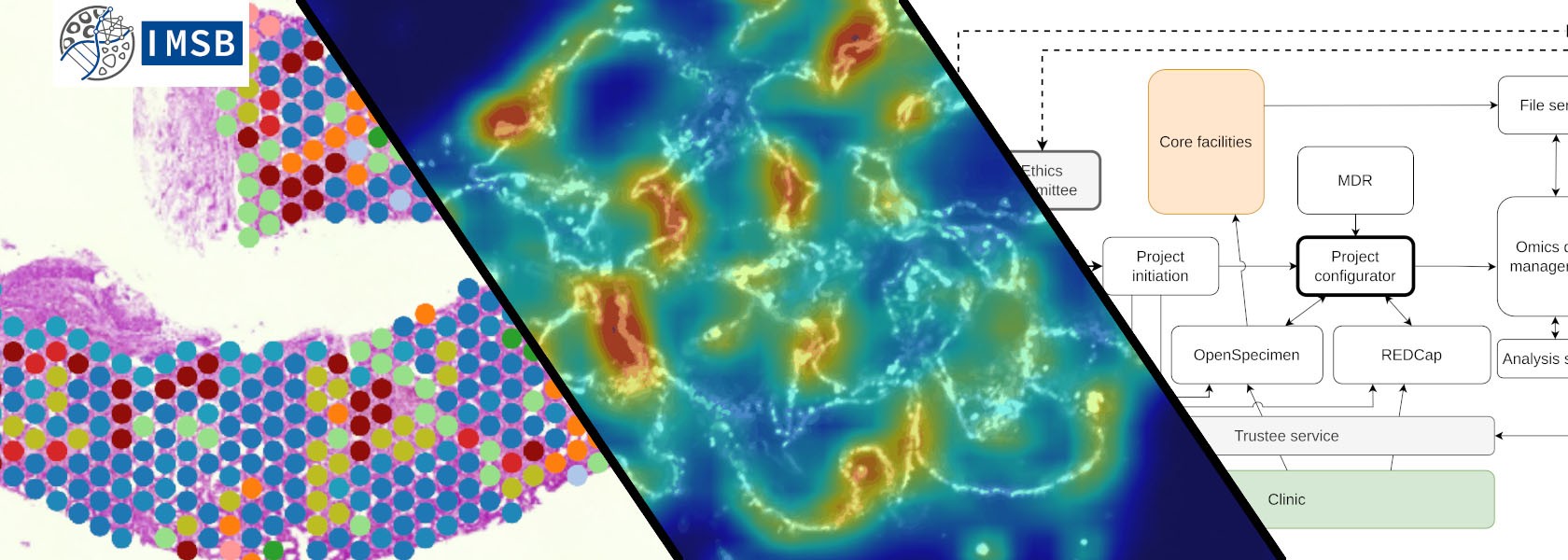
We strive to deepen the understanding of human pathology through computational methodologies and ultimately improve patient outcomes through innovative clinical decision support systems and novel therapeutic approaches. We maintain close collaboration with the medical clinics of the UKE to ensure our research remains directly applicable to clinical practice. Our approach integrates and analyzes vast amounts of biomedical data using cutting-edge technologies such as multi-omics analyses, drug-target interaction, deep learning or spatial transcriptomics.
Our commitment to advancing scientific knowledge extends beyond our research efforts. We offer courses in machine and deep learning to foster interdisciplinary collaboration among physicists, computer scientists, and medical professionals. Additionally, our research groups focus on specialized areas such as genomic AI, computational pathology, biomedical data analysis, and data integration.
Current projects – Bonn Lab
MAXOMOD
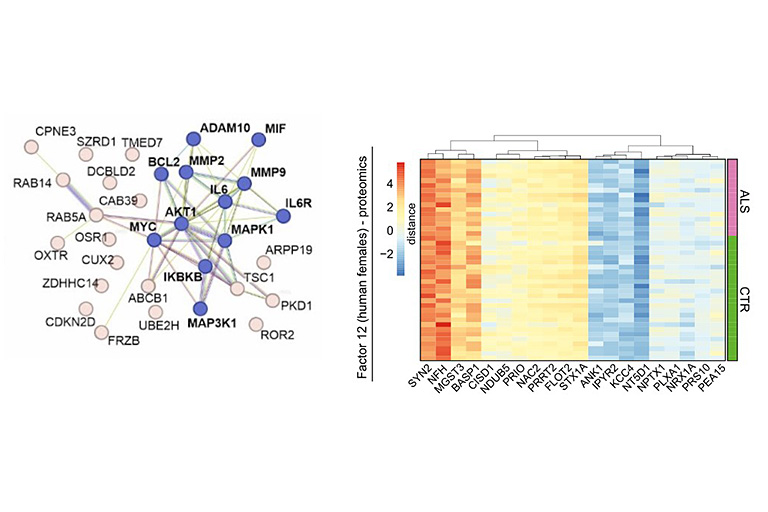
Motor neuron diseases such as amyotrophic lateral sclerosis (ALS) are rare neurodegenerative diseases with poor prognosis and insufficient treatment options. Patients with ALS develop increasing muscle weakness as the disease progresses, ultimately leading to death when lung function is lost. Existing therapy options only treat the symptoms of the disease and can hardly influence the course of neurodegeneration. Therefore, there is a need for research to ensure that new therapies arrive in the treatment of motor neuron disease. The goal of the project is to develop new therapeutic strategies as well as biomarkers that enable early diagnosis of the disease. To this end, new molecular targets are to be identified that could serve as potential targets for drug therapies.
Since RNA metabolism plays a crucial role in the pathogenesis of ALS, a protein-based approach will not be sufficient to comprehensively characterize disease-relevant pathways. We will thus combine multiple analytic methods from genomics, transcriptomics and microRNAomics up to proteomics, phosphoproteomics and metabolomics followed by multivariate semantic data integration to identify new disease-relevant pathways and biomarkers related to axono-synaptic pathology.
PCAI
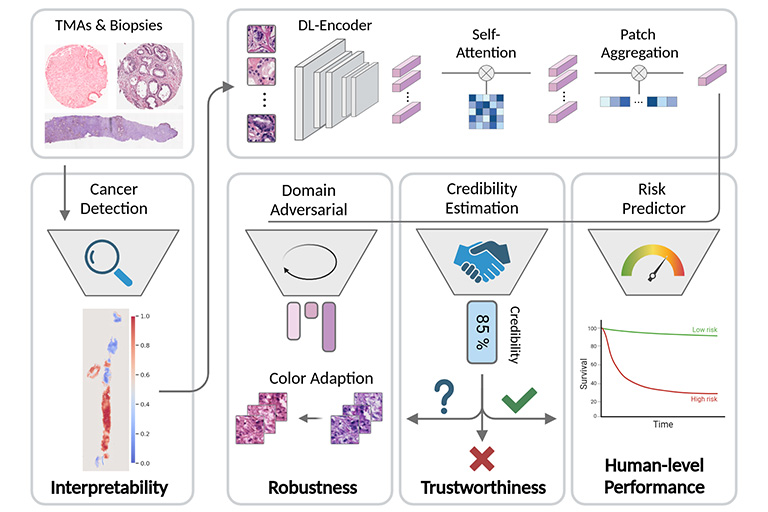
In clinical practice, the diagnosis of Prostate cancer (PCa) and the treatment decision are based on Gleason scores and nomograms. However, Gleason scores suffer from high inter-observer variability and nomograms are only able to model linear dependencies between patient parameters. Therefore, the goal of our research is to provide individual and objective prognoses for PCa patients through predicting relapse after radical prostatectomy (RPE). To this end we build and train deep learning-based models that predict the probability of a patient having a relapse from either H&E-stained images of biopsies after the RPE or electronic health records that reflect the patient’s clinical history related to prostate cancer.
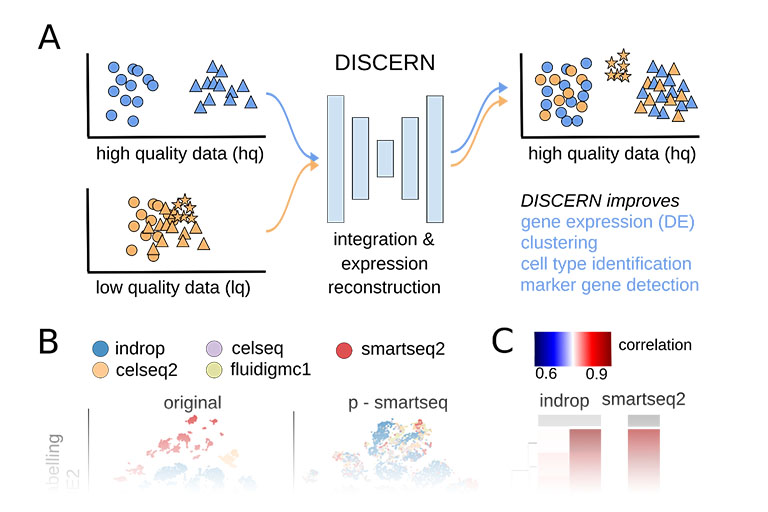
DISCERN
Single cell sequencing provides detailed insights into biological processes including cell differentiation and identity. While providing deep cell-specific information, the method suffers from technical constraints, most notably a limited number of expressed genes per cell, which leads to suboptimal clustering and cell type identification. We developed DISCERN, a novel deep generative neural network that reconstructs missing single cell gene expression using a reference dataset. DISCERN based expression inference results in greatly improved cell clustering, cell type and activity detection, and can lead insights into the cellular regulation of disease. For example, we used DISCERN to detect two cell types with a potential role in adverse COVID-19 outcome. DISCERN can be easily integrated into existing single cell sequencing workflows and readily adapted to enhance various other biomedical data types.
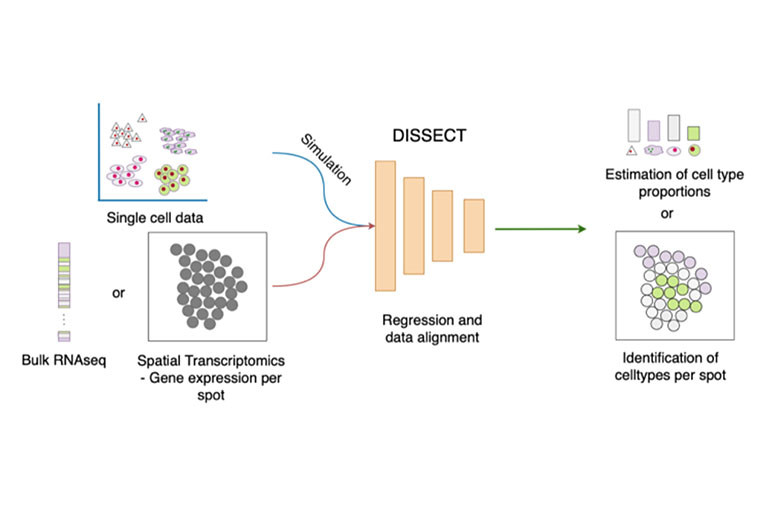
DISSECT
Bulk RNA-seq has enabled contributions in biomarker discovery, survival prediction and optimizing potential therapeutic targets. In comparison to single cell RNA-seq, the method is affordable and as such larger cohorts can be studied. While providing unprecedented insights at tissue level, the method is unsuitable to study cell type specific changes. Previously, IMSB developed SCADEN, a novel deep neural network ensemble framework that enabled accurate transfer of cell type specific knowledge from single cell RNA-seq to recover cell type landscape of bulk RNA-seq. For example, SCADEN revealed an increasing vascularization in Alzheimer’s disease. However single cell datasets suffer from dropouts and they differ in their scope from one study to another. Therefore, we developed DISSECT that utilizes information from bulk RNA-seq directly to estimate more accurate and robust cell type proportions. For example, we used DISSECT to recover cell type proportions of over 20 immune cell types in colons from individuals with inflammatory bowel disease where we identified an increasing infiltration of immune cells as well as a significantly increased number of activated resident cells. DISSECT is highly scalable and is easily integratable in existing bioinformatics workflows.
Team – Bonn Lab


Prof. Dr. med. Stefan Bonn
Head of lab - Systems Biology
E-mail address:






















Key Publications – Bonn Lab
Halder, R., Hennion, M., Vidal, R. O., Shomroni, O., Rahman, R. U., Rajput, A., ... & Bonn, S. (2016). DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nature neuroscience, 19(1), 102-110.
Marouf, M., Machart, P., Bansal, V., Kilian, C., Magruder, D. S., Krebs, C. F., & Bonn, S. (2020). Realistic in silico generation and augmentation of single-cell RNA-seq data using generative adversarial networks. Nature communications, 11(1), 166.
Menden, K., Marouf, M., Oller, S., Dalmia, A., Magruder, D. S., Kloiber, K., ... & Bonn, S. (2020). Deep learning–based cell composition analysis from tissue expression profiles. Science advances, 6(30), eaba2619.
Hausmann, F., Ergen, C., Khatri, R., Marouf, M., Hänzelmann, S., Gagliani, N., ... & Bonn, S. (2023). DISCERN: deep single-cell expression reconstruction for improved cell clustering and cell subtype and state detection. Genome Biology, 24(1), 212.
Alumni - PhD | MD
- Ashish Rajput
– Usinf cell type-specific methods to understand molecular process in the brain – 11/2013 - 04/2018, Degree obtained "Dr. rer. nat." - Anna-Maria Liebhoff
– Detection of pathogenic infections in neurological disorders through recycling of gene expression data – 09/2017 - 02/2020, Degree obtained "Dr. rer. nat." - Daniel Sumner Magruder
– Development of interactive software an AT-based algorithms fort he analysis of biomedical data – 09/2017 - 10/2021, Degree obtained "Dr. rer. biol. hum." - Martin Klaus
– Development of new tools to analyze kidney biopsies based on deep learning – 2021, Degree obtained "Dr. med." - Robert Gaudin
– Entwicklung eines Deep learning Algorithmus zur Detektion von periapikalen Aufhellungen in Panoramaschichtaufnahmen – 2021, Degree obtained "Dr. med." - Anupriya Dalmia
– Elucidating the Genetic Landscape oft he Frontotemporal Dementias using Next-Generation Sequencing and In-Silico Analyses – 11/2017 - 04/2021, Degree obtained "Dr. rer. nat." - Anastasia Illarionova
– Identification of genetic risk factors for Parkonson´s disease – 09/2018 - 02/2023, Degree obtained "Dr. rer. nat." - Fabian Hausmann
– Applications of deep generative modeling approaches on Omics data – 01/2020 - 03/2023, Degree obtained "Dr. rer. nat." - Esther Dietrich
– Enabling digital pathology for prostate cancer stratification – 01/2020 - 12/2022, Degree obtained "Dr. rer. nat." - Yu Zhao
– Computational characterization of T cells in inflammatory disease – 01/2018 - 05/2022, Degree obtained "Dr. rer. nat."