 Research at the UKE?
Research at the UKE?
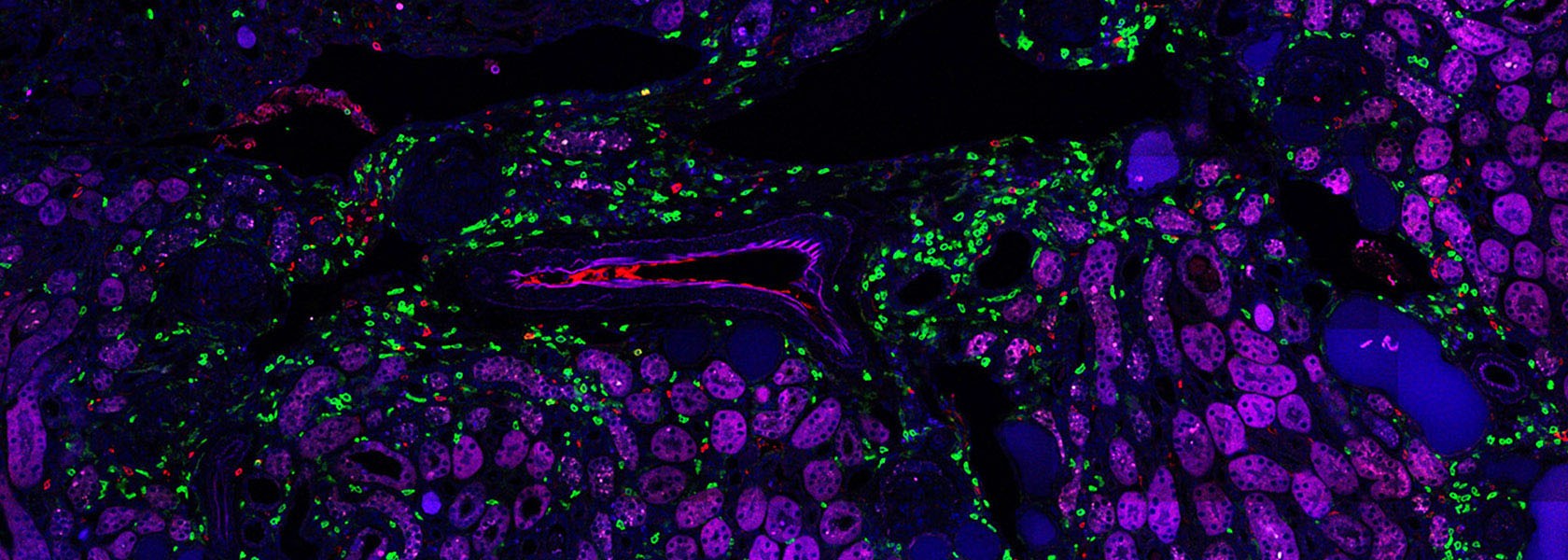
Panzer and Krebs Lab – Translational Immunology
Immune-mediated kidney diseases are showing an increasing prevalence. Yet, their diagnosis is often complex and limited therapies are available. Unravelling the site-specific immunity can lead to improved diagnostics and targeted therapies, addressing the significant impact these diseases have on patients' lives and potentially leading to personalized treatment approaches.

“Improving the understanding of tissue-specific immunity in the kidney”
Prof. Dr. Ulf Panzer
/ck_final_3.jpg)
"Targeting pro-inflammatory T cell responses in renal autoimmunity"
Prof. Dr. Christian Krebs
Project details and goals
CD4+ T cells are crucial in autoimmune and chronic inflammatory diseases by coordinating the immune response. Characterizing specific cytokine-producing CD4+ T cell subsets has enhanced our understanding of organ-specific immunity mechanisms. However, many aspects of T cell behavior and regulation in kidney damage remain unclear.
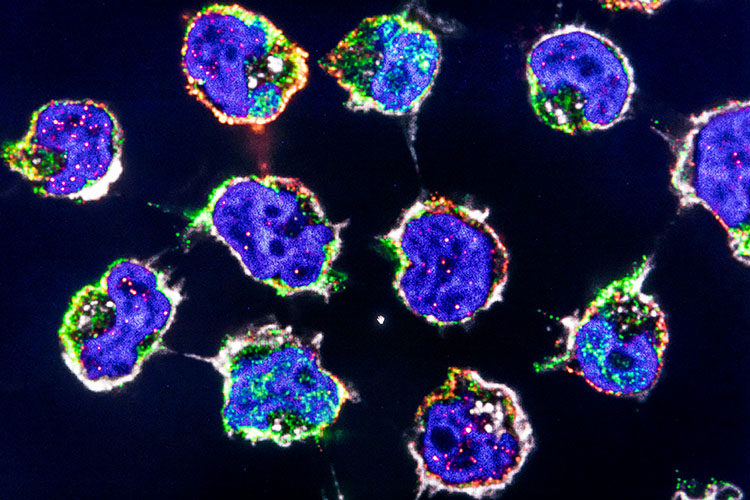
To address this gap, we will use multiomics approaches to:
- investigate T cell migration mechanisms, including retention and emigration;
- study Type I and Type III immune responses and their associated receptors and chemokines in kidney diseases;
- conduct a high-dimensional analysis of CD4+ T cell responses through systems immunological methods.
In summary, our research aims to deeply understand the CD4+ T cell chemokine network and its role in immunomediated glomerulonephritis, laying the groundwork for pathogenesis-based anti-cytokine treatments or specific depletion of pathogenic renal T cells.
Current projects – Panzer Lab

Mechanisms and function of CD4+ T cell trafficking in crescentic GN
CD4+ T cells are pivotal in orchestrating immune responses, including in autoimmune conditions like crescentic glomerulonephritis (cGN). The migration of these T cells to target organs and their effector functions are crucial in both protective and pathological responses. Understanding the dynamics of T cell subsets, such as TH1, TH17, and Treg cells, in the kidney is essential for elucidating disease mechanisms. Experimental models reveal that T cell infiltration, rather than proliferation or apoptosis, primarily drives renal inflammation in cGN. Investigating T cell emigration from the kidney via lymphatic vessels will provide insights into resolving or perpetuating local immune reactions, impacting tissue-resident memory T cell function. The "Kaede mouse system" offers a promising tool to study T cell migration in the kidney and its implications for immune regulation.
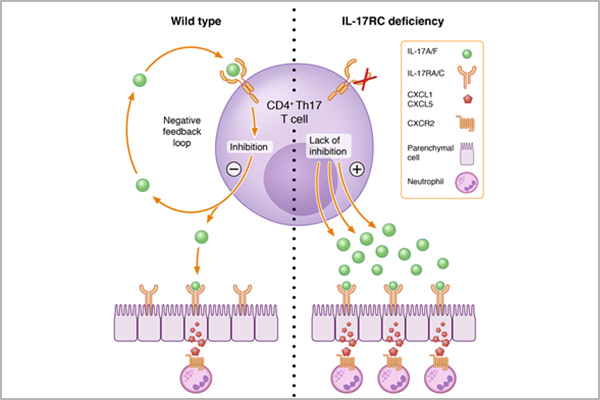
Renal CD4+ T cell cytokine network – Translational regulation and function of IL-17 cytokine mRNAs
The Th17/IL-17 pathway is implicated in tissue-specific immunity, particularly in immune-mediated kidney diseases. Despite advancements, understanding the regulation and biological functions of IL-17 cytokines and their receptors remains incomplete. Research shows that CD4+ tissue-resident memory T cells (TRM cells) express proinflammatory cytokine mRNAs without causing inflammation under normal conditions but may contribute to pathology in glomerular diseases upon stimulation. Further investigation revealed that the integrated stress response (ISR) / eukaryotic translation initiation factor 2α (eIF2α) pathway regulates cytokine production in CD4+ TRM cells, with activation of ISR/eIF2α under homeostasis storing cytokine mRNA and preventing inflammation, while under inflammatory conditions, dephosphorylation of eIF2α leads to immediate cytokine translation, suggesting tight control of TRM cell effector functions.

CD4+ T cell subsets and their signature cytokines in crescentic GN patients
The understanding of pathogenic CD4+ T-cell-driven immune responses, particularly Th1 and Th17, in crescentic glomerulonephritis (GN) largely stems from animal models, with limited analysis in patients. Identifying disease-promoting CD4+ T-cell populations and cytokines in crescentic GN patients remains incomplete but is crucial for translating findings to clinical practice, as seen in other immune-mediated diseases like psoriasis and rheumatoid arthritis. Collaborative efforts have established platforms for analyzing T-cell subsets in renal tissue injury, aiming to decode the CD4+ T-cell immune signature in ANCA-GN patients using single-cell and spatial transciptomic techniques. This interdisciplinary approach involves nephrology, immunology, and systems biology, relying on clinical data and multi-OMIC analysis to generate a comprehensive CD4+ T-cell immune signature atlas, which could inform pathogenesis-based treatment strategies.
Current projects – Krebs Lab

Crosse-tissue T cell clonality and role of intestinal microbiota from patients with glomerulonephritis
Interorgan homeostasis and tissue crosstalk is a complex, yet curial system for immune cell function. Here, we aim to investigate the interorgan relationship of T cells in patients with glomerulonephritis (GN) by analyzing T cell receptor sequences and RNA expression profiles in peripheral blood, kidney, lung, and colon tissues. Preliminary results show the feasibility of identifying developmental trajectories of renal T cells. Additionally, the we plan to assess the composition of intestinal microbiota and metabolites in patients with ANCA-GN and healthy controls using metagenomic sequencing and metabolomics. Bioinformatics analysis will integrate single-cell RNA sequencing data with microbiota and metabolite data to identify connections between T cell populations, microbial metabolites, and signaling pathways, facilitating a comprehensive understanding of immune-microbiota interactions in GN

Regulation of Th17 cell tissue adaption
The Th17 immune response is a key driver of immune mediated kidney disease. Here, we aim to investigate the role of transcription factors in regulating Th17 cell phenotypes in experimental glomerulonephritis (GN), particularly focusing on identifying factors contributing to pathogenic and regulatory stages of Th17 cells. We plan to generate Th17 cells deficient for specific transcription factors and utilize a pooled screening approach to further investigate transcription factors and intracellular pathways. Additionally, we will investigate the role of selected candidate genes potentially involved in tissue adaptation using a CRISPR/Cas9 approach in mouse models of GN and colitis, aiming to narrow down promising candidates for further functional testing. The iCROP-seq screening approach will allow us to assess the impact of different genes on tissue-specific T cell adaptation, particularly focusing on transcription factors upregulated in Th17 cells specific to kidney or shared among different tissues.

Deciphering the role of the intestinal microbiota in impacting Th17 cell tissue-adaptation
Germ-free mice and antibiotic-treated mice show reduced Th17 cells in the kidney and are protected against nephritis development, suggesting the crucial role of the intestinal microbiota. We collected stool samples from ANCA-GN patients and healthy controls to investigate the microbiota's composition and microbial metabolites. Based on these analyses we will induce this microbiota signature in germ-free mice. The impact of microbiota transfer on renal T cell responses will be analyzed. Furthermore, patient-specific gnotobiotic mice and oligoclonal mice will be investigated to overcome TCR repertoire variations. Antibiotic treatment-induced T cell modulation in the intestine will be studied, assessing its effect on disease severity, microbiota, metabolites, and T cell plasticity in both intestine and kidney.
Team – Panzer Lab


Prof. Dr. med. Ulf Panzer
Head
E-mail address:











Team – Krebs Lab


Prof. Dr. med. Christian Krebs
Head
E-mail address:







Key Publications – Panzer Krebs Lab
Jonas Engesser, Robin Khatri, Darius P Schaub, Yu Zhao , Hans-Joachim Paust, Zeba Sultana, Nariaki Asada, Jan-Hendrik Riedel , Varshi Sivayoganathan, Anett Peters, Anna Kaffke, Saskia-Larissa Jauch-Speer, Thiago Goldbeck-Strieder, Victor G Puelles, Ulrich O Wenzel, Oliver M Steinmetz, Elion Hoxha, Jan-Eric Turner, Hans-Willi Mittrücker, Thorsten Wiech, Tobias B Huber, Stefan Bonn, Christian F Krebs, Ulf Panzer
PMID: 39300109 PMCID: PMC11413367 DOI:
Abstract
Antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis is a life-threatening autoimmune disease that often results in kidney failure caused by crescentic glomerulonephritis (GN). To date, treatment of most patients with ANCA-GN relies on non-specific immunosuppressive agents, which may have serious adverse effects and be only partially effective. Here, using spatial and single-cell transcriptome analysis, we characterize inflammatory niches in kidney samples from 34 patients with ANCA-GN and identify proinflammatory, cytokine-producing CD4+ and CD8+ T cells as a pathogenic signature. We then utilize these transcriptomic profiles for digital pharmacology and identify ustekinumab, a monoclonal antibody targeting IL-12 and IL-23, as the strongest therapeutic drug to use. Moreover, four patients with relapsing ANCA-GN are treated with ustekinumab in combination with low-dose cyclophosphamide and steroids, with ustekinumab given subcutaneously (90 mg) at weeks 0, 4, 12, and 24. Patients are followed up for 26 weeks to find this treatment well-tolerated and inducing clinical responses, including improved kidney function and Birmingham Vasculitis Activity Score, in all ANCA-GN patients. Our findings thus suggest that targeting of pathogenic T cells in ANCA-GN patients with ustekinumab might represent a potential approach and warrants further investigation in clinical trials.
Leon U B Enk, Malte Hellmig, Kristoffer Riecken, Christoph Kilian, Paul Datlinger, Saskia L Jauch-Speer, Tobias Neben, Zeba Sultana, Varshi Sivayoganathan, Alina Borchers, Hans-Joachim Paust, Yu Zhao, Nariaki Asada, Shuya Liu, Theodora Agalioti, Penelope Pelczar, Thorsten Wiech, Christoph Bock, Tobias B Huber, Samuel Huber, Stefan Bonn, Nicola Gagliani, Boris Fehse, Ulf Panzer, Christian F Krebs
DOI:
Abstract
Pro-inflammatory CD4+ T cells are major drivers of autoimmune diseases, yet therapies modulating T cell phenotypes to promote an anti-inflammatory state are lacking. Here, we identify T helper 17 (TH17) cell plasticity in the kidneys of patients with antineutrophil cytoplasmic antibody-associated glomerulonephritis on the basis of single-cell (sc) T cell receptor analysis and scRNA velocity. To uncover molecules driving T cell polarization and plasticity, we established an in vivo pooled scCRISPR droplet sequencing (iCROP-seq) screen and applied it to mouse models of glomerulonephritis and colitis. CRISPR-based gene targeting in TH17 cells could be ranked according to the resulting transcriptional perturbations, and polarization biases into T helper 1 (TH1) and regulatory T cells could be quantified. Furthermore, we show that iCROP-seq can facilitate the identification of therapeutic targets by efficient functional stratification of genes and pathways in a disease- and tissue-specific manner. These findings uncover TH17 to TH1 cell plasticity in the human kidney in the context of renal autoimmunity.
Paust HJ, Song N, De Feo D, Asada N, Tuzlak S, Zhao Y, Riedel JH, Hellmig M, Sivayoganathan A, Peters A, Kaffke A, Borchers A, Wenzel UO, Steinmetz OM, Tiegs G, Meister E, Mack M, Kurts C, von Vietinghoff S, Lindenmeyer MT, Hoxha E, Stahl RAK, Huber TB, Bonn S, Meyer-Schwesinger C, Wiech T, Turner JE, Becher B, Krebs CF, Panzer U. CD4+ T cells produce GM-CSF and drive immune-mediated glomerular disease by licensing monocyte-derived cells to produce MMP12. Sci Transl Med. 2023 Mar 15;15(687):eadd6137. doi:
Abstract
GM-CSF in glomerulonephritisDespite glomerulonephritis being an immune-mediated disease, the contributions of individual immune cell types are not clear. To address this gap in knowledge, Paust et al. characterized pathological immune cells in samples from patients with glomerulonephritis and in samples from mice with the disease. The authors found that CD4+ T cells producing granulocyte-macrophage colony-stimulating factor (GM-CSF) licensed monocytes to promote disease by producing matrix metalloproteinase 12 and disrupting the glomerular basement membrane. Targeting GM-CSF to inhibit this axis reduced disease severity in mice, implicating this cytokine as a potential therapeutic target for patients with glomerulonephritis. -CM.
(DOI:
Zhao Y, Kilian C, Turner JE, Bosurgi L, Roedl K, Bartsch P, Gnirck AC, Cortesi F, Schultheiß C, Hellmig M, Enk LUB, Hausmann F, Borchers A, Wong MN, Paust HJ, Siracusa F, Scheibel N, Herrmann M, Rosati E, Bacher P, Kylies D, Jarczak D, Lütgehetmann M, Pfefferle S, Steurer S, Zur-Wiesch JS, Puelles VG, Sperhake JP, Addo MM, Lohse AW, Binder M, Huber S, Huber TB, Kluge S, Bonn S, Panzer U, Gagliani N, Krebs CF. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing GM-CSF in the lungs of severe COVID-19 patients. Sci Immunol. 2021 Feb 23;6(56):eabf6692. doi:
(DOI:
Krebs CF, Reimers D, Zhao Y, Paust HJ, Bartsch P, Nuñez S, Rosemblatt MV, Hellmig M, Kilian C, Borchers A, Enk LUB, Zinke M, Becker M, Schmid J, Klinge S, Wong MN, Puelles VG, Schmidt C, Bertram T, Stumpf N, Hoxha E, Meyer-Schwesinger C, Lindenmeyer MT, Cohen CD, Rink M, Kurts C, Franzenburg S, Koch-Nolte F, Turner JE, Riedel JH, Huber S, Gagliani N, Huber TB, Wiech T, Rohde H, Bono MR, Bonn S, Panzer U, Mittrücker HW. Pathogen-induced tissue-resident memory TH17 (TRM17) cells amplify autoimmune kidney disease. Sci Immunol. 2020 Aug 7;5(50):eaba4163. doi:
(DOI:
Alumni - PhD
- Zhao, Yu (Postdoctoral Research Fellow at Harvard Medical School)
– High dimensional renal CD4+ T cell analysis in human ANCA-GN patients – 2018-2021
Alumni - MD
- Robben, Lennart
– Regulation of the CD4+ T cell reponse by steroids in autoimmune kidney disease – 2019-2020 - Ginsberg, Pauline
– Bedeutung des “Integrated stress response (IRS)” in der Regulation von Zytokinen gewebsspezifischer T Gedächtniszellen 2020-2021 - Kleiner, Jens
– Functional Role of Tbet-dependent Th17 to Th1 cell plasticity in immune-mediated diseases – 2020-2021