1. Molecular mechanisms of integrin trafficking
Numerous diseases such as cancer progression, inflammation and degenerative disorders are results of deregulated cell adhesion and migration. The integrin family of transmembrane cell surface receptors have a decisive impact on the migratory and invasive capacities of cancer and immune cells. Furthermore, a variety of pathogens use integrins to invade host cells for infection, extending the relevance of integrins to infectious diseases. It is becoming increasingly evident that not only activation/deactivation, but also the endo-/exocytic cycle of integrins plays an important role in mediating integrin function. The small Ras-family GTP-binding protein Rab21 is a major regulator in the endocytic trafficking of integrins. However, on a molecular or structural level the mode of action remains poorly understood mainly due to the lack of known Rab21-interacting proteins. The aim of our research is to gain insights how Rab21 organises integrin trafficking on a structural-mechanistic level
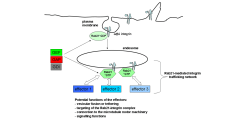
(Figure 1).
The proposed Rab21-regulated integrin trafficking complex.
Rab21 might bind the cargo integrin heterodimer and downstream effectors in concert allowing recruiting several effectors within a potential integrin-Rab21 cluster.
We aim to identify the downstream effectors and examine how Rab21 organises the trafficking of integrins with these effectors. To identify the Rab21-integrin trafficking network, three sub-projects are ongoing or will be implemented in the near future.
A). Identification of Rab21 interacting proteins involved in integrin trafficking.
Novel Rab21 upstream regulators (GAPs and GEFs) or downstream effector proteins are identified using a quantitative mass spectrometry-based protein-protein interaction screen and tested regarding their influence on integrin trafficking
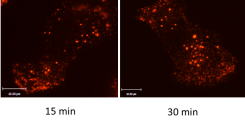
(Figure 2).
Visualisation of integrin endocytosis.
Cell surface #1-Integrins on MDA MB 231 breast cancer cells are labelled with an anti-integrin antibody and internalised. After removal of residual surface-bound antibodies, an increase of endocytosed integrins by time can be visualised using confocal microscopy.
B). Structural analysis of the Rab21-integrin trafficking network.
After their identification, Rab21 interaction partners are crystallised in complexes with Rab21 to obtain a structural snapshot of the Rab21-regulated integrin trafficking network. The X-ray structures will provide insights into the function of the trafficking complex and answer the question how Rab21 manages to transport cargo (integrin) and bind effector proteins in parallel.
C). Functional analysis of the Rab21 interacting proteins.
We will investigate the biochemical and cell biological function of the identified proteins and address the question how these proteins affect the dynamics of integrin endocytosis. Different microscopy techniques (confocal microscopy, live cell imaging, TIRF) can be employed.
In our projects, we employ a multidisciplinary approach starting with a biochemical screen and combining various state of the art and cutting edge cell and structural biological tools to maximise our mechanistic understanding of the question how integrins are trafficking from the plasma membrane to endosomes and how this trafficking influences different disease processes.
The project is funded by the DFG (VE 750/2-1).
2. Time-resolved crystallisation of an enzymatic GAP-GTPase complex
"Seeing is believing" - a key to understand biological molecules is to identify their 3D-structure mainly using X-ray crystallography. However, one problem of this method is that it shows only a frozen image of a protein or a protein complex. To understand enzymatic reactions in great detail, it is crucial to know the time-ordered sequence of events of the biochemical reaction. Novel methods in structure determination employ time-resolved X-ray crystallographic methods to image single steps during the enzymatic reaction creating a "molecular movie"
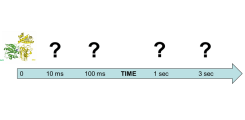
(Figure 3).
Scheme of a molecular movie from an enzymatic GTPase-GAP reaction.
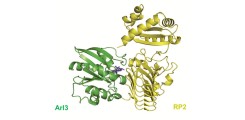
The structure of the ground-state of an enzymatic reaction (here the GTPase-GAP system: Arl3-RP2) can be determined by classical X-ray structure determination methods. To obtain structural snapshots during the reaction, we apply novel time-resolved methods.
Time-resolved methods exist, but, due to both instrument availability and sensitive sample requirements, they have not been widely applied to macromolecular systems, especially for time resolutions below one second. Laue diffraction has been the main technique of choice to obtain time-resolved structural information. Nevertheless the pump-probe Laue method is highly demanding not only on the technical setup, but also on the quality of the crystal samples.
With this research project we try to establish a proof-of-principle set-up to combine the pump-probe Laue method with the approach of serial femtosecond crystallography (SFX) on macromolecular systems that can be triggered with caged compounds. Caged compounds are light-sensitive probes that keep the enzymatic complex in the crystal in an inactive form. UV-irradiation liberates the trapped molecule to induce the reaction start inside the crystal. For the structural determination of the kinetics of enzymatic reactions we will focus on the small GTPase Arl3 and its co-enzyme, the GTPase-activating protein (GAP) RP2, a system important in the hereditary eye disease Retinitis pigmentosa. The GTPase mechanism is of very high, general importance in cell biology with particular impact on disease processes, especially cancer and infectious diseases.
The project is done in close collaboration with PD Dr. Markus Perbandt from the DESY and funded by the "PIER Ideenfond" from the University of Hamburg and the DESY.
Staff Group Veltel